Link to publication: https://www.cell.com/stem-cell-reports/fulltext/S2213-6711(22)00423-4
In our most recent work in collaboration with colleagues from Life & Brain and the University of Bonn we show how the combination of many image-derived quantitative features, and the use of machine learning (ML) can be useful for in vitro Parkinson’s disease modelling using human neurons.
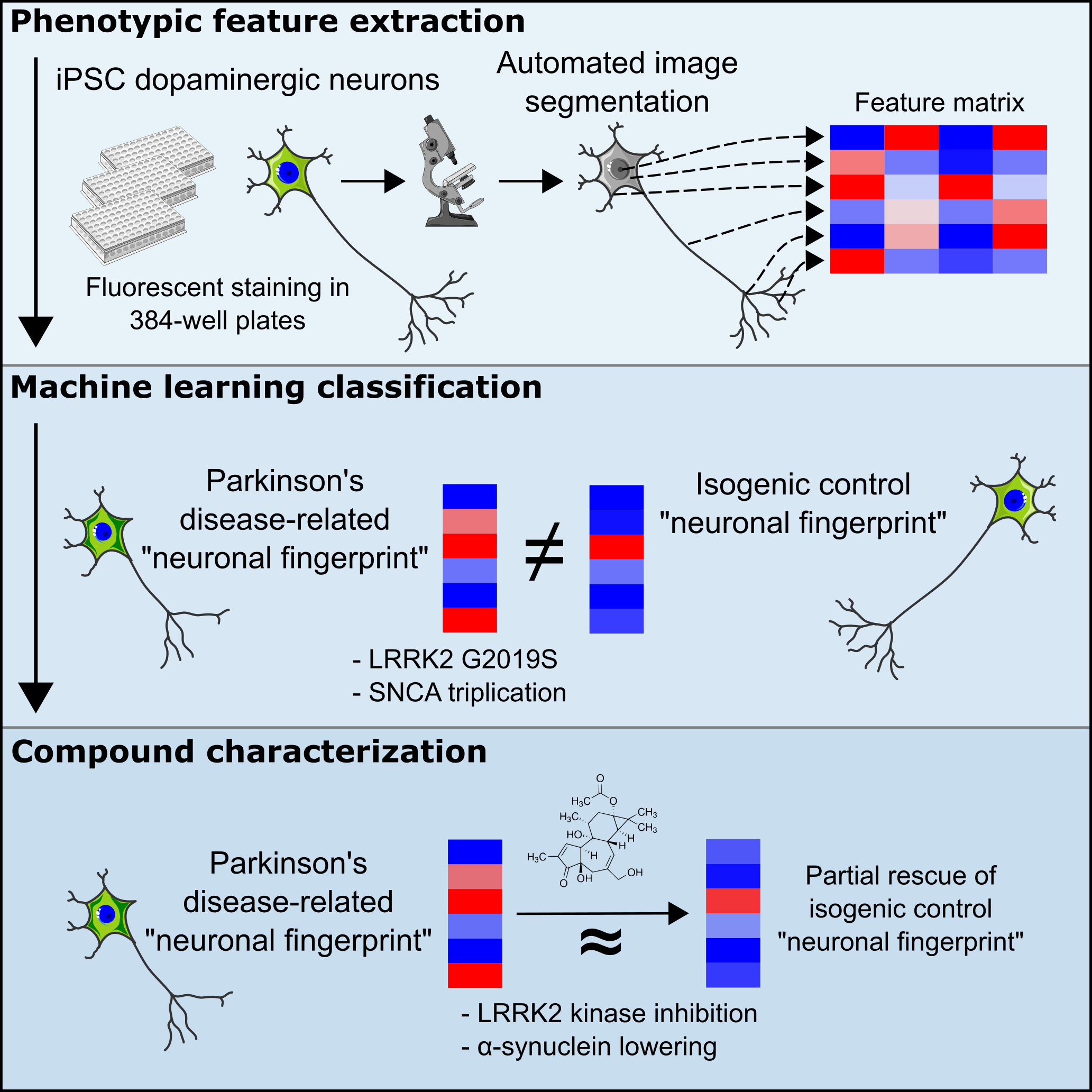
We started out with the idea that Parkinson’s Disease-specific signatures might be detectable based on the vast information digital images offer to us. Often researchers focus on a single phenotype only. We wanted to take many into account. We used two Parkinson’s Disease patient-derived iPSC models and the corresponding isogenic controls with repaired mutation. The first model carried a LRRK2 G2019S mutation, while in the second model the SNCA gene was triplicated. Both mutations have strong links with Parkinson’s disease. All cell lines were differentiated into midbrain dopaminergic neurons, the neuron type most vulnerable in Parkinson’s disease. The neurons were then stained, imaged and data was extracted from images. All steps were automated and are suitable for up-scaling. 126 morphological features covering intensities, correlations, shapes, textures, and other image-quantifiable elements were extracted. An additional 54 mitochondrial features were extracted using a separate staining panel. Next, we used supervised binary machine learning classifiers to group known and unknown classes into categories. The idea was that image features would be sufficient to classify neurons based on genotype or chemical compound treatment.
First, we focused on genotype characterization. Our multi-feature approach classified neurons correctly using two different ML algorithms. “Unseen” healthy neurons classified similarly as isogenic controls. From a phenotypic perspective, “healthy” neurons therefore seem to be similar to each other. Secondly, we focused on chemical compounds and asked the question: Can chemical modulators shift a “Parkinson’s Disease” phenotypic profile towards a phenotypic profile more similar to “healthy cells”? We evaluated multiple molecules: Three LRRK2 kinase inhibitors and PEP005 and Prostratin, two molecules known to lower α-synuclein levels in cell culture. Two of the three LRRK2 kinase inhibitors shifted the “Parkinson’s Disease” phenotypic profile towards a “healthy” phenotypic profile. The rescue was partial because not all features were affected alike, highlighting the value of our approach to identifying features of interest. The same effect was observed when we included mitochondria-derived phenotypic features. Additionally, we tested PEP005 and Prostratin, both α-synuclein lowering compounds, and checked their effect on the phenotypic profiles of control and LRRK2 mutant neurons. Both molecules led to small, but detectable shifts toward a “healthy” profile. PEP005 and Prostratin effects became more pronounced when we slightly changed the staining panel and included the lysosomal marker LAMP2. Both molecules are assumed to act via the lysosome. To check whether chemical compound modulation of phenotypic profiles would also work in a completely different mutation background we repeated our experiments in dopaminergic neurons derived from an isogenic SNCA triplication model and obtained similar results. Finally, we showed in both models that phenotypic profiles are comparable after 37 and 50 days of differentiation, highlighting the stability of phenotypic profiles over time.
In summary, we show that combining many subtle, but biological meaningful phenotypes, improves the predictive value of otherwise variable dopaminergic neuron models, for example for chemical compound characterization and eventually implementation in the drug discovery process.